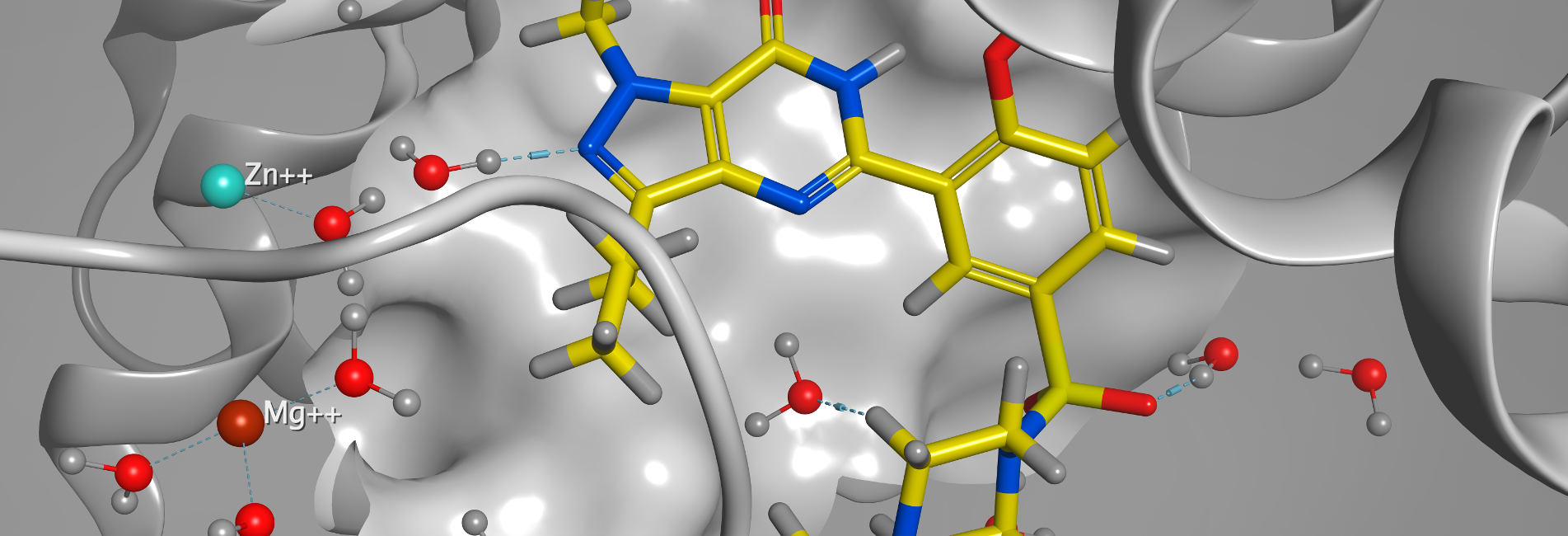
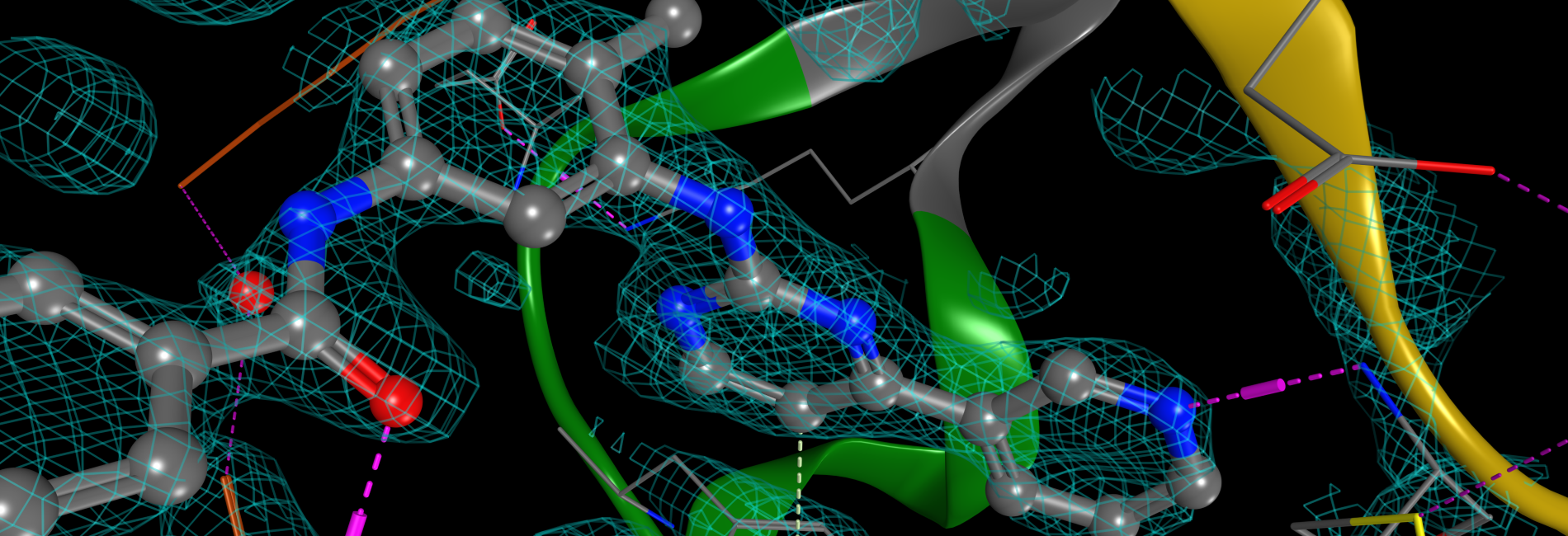
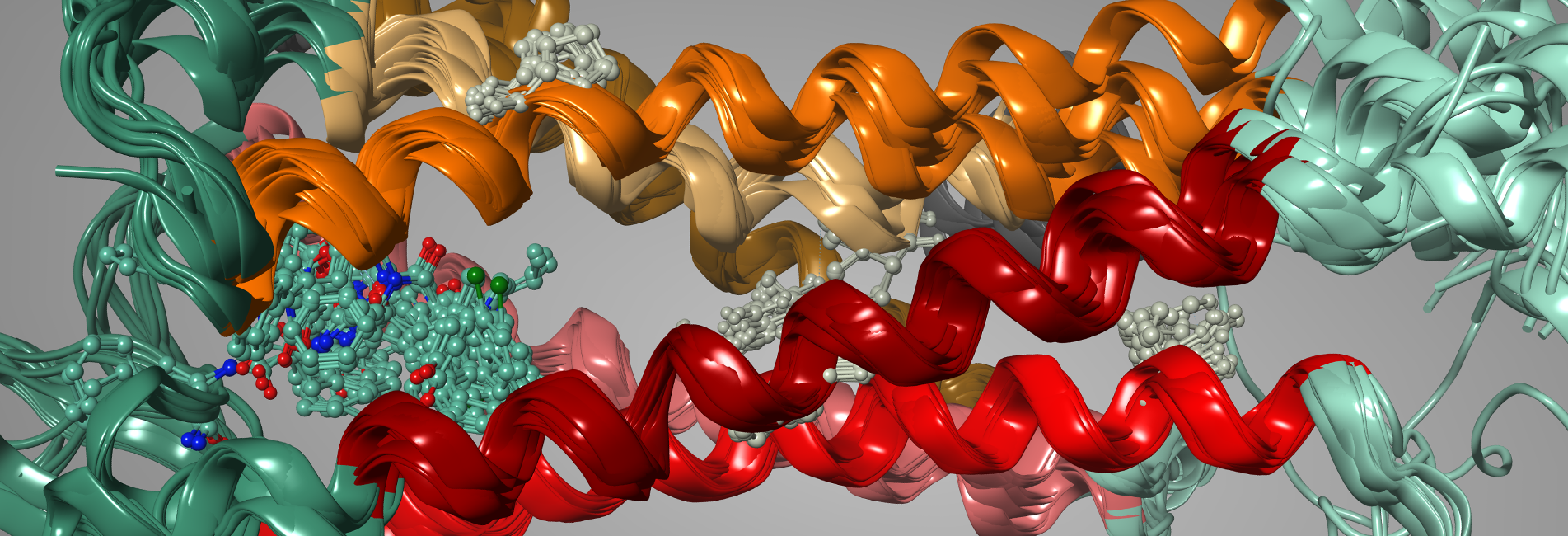
COMPUTER-AIDED MOLECULAR DESIGN
CCG is a leading developer and provider of Molecular Modeling, Simulations and Machine Learning software to Pharmaceutical and Biotechnology companies as well as Academic institutions throughout the world. CCG continuously develops new technologies with its team of mathematicians, scientists and software engineers and through scientific collaborations with customers.
Upcoming Events
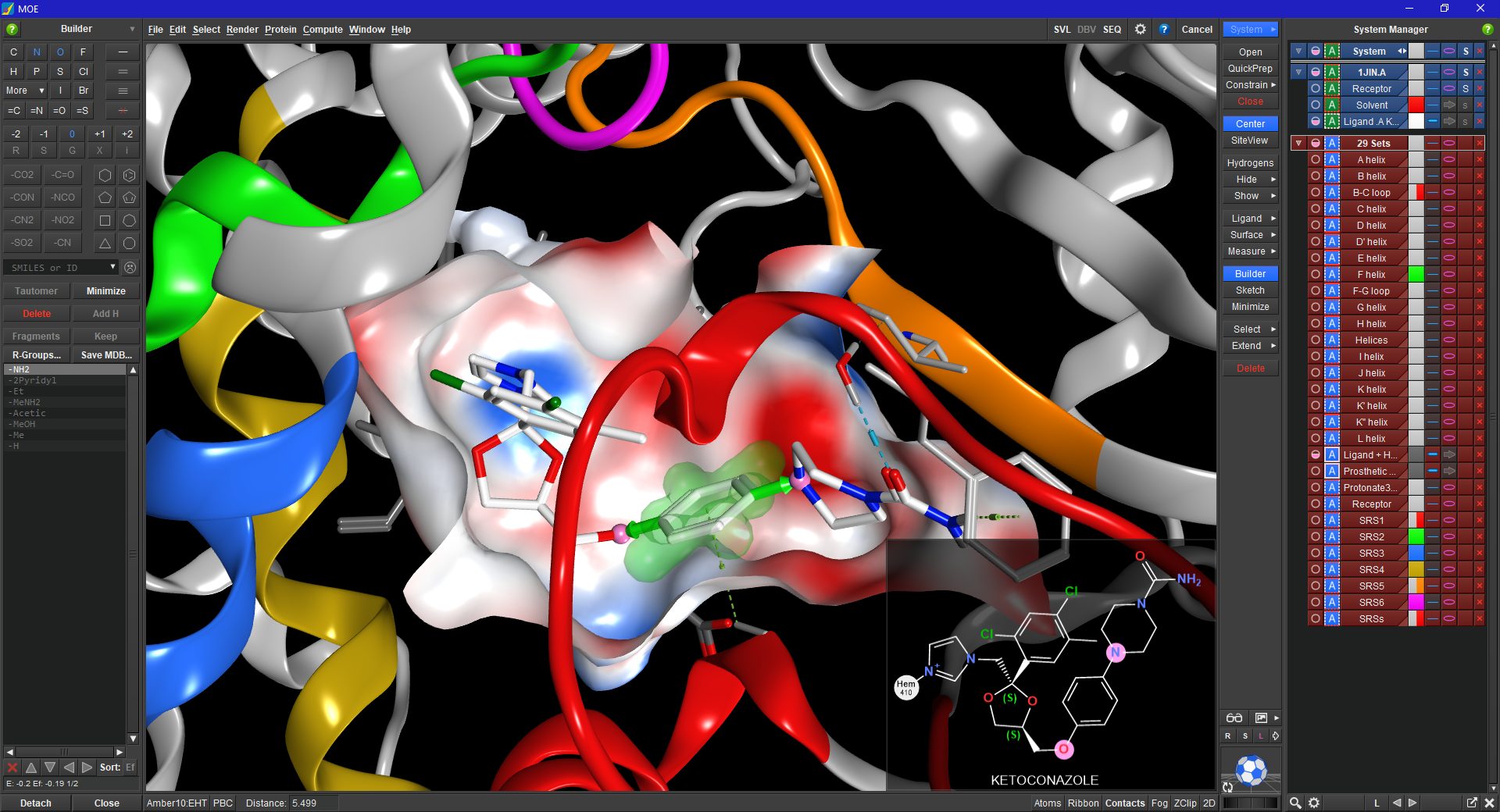
MOE 2022.02
Latest Features
On-the-fly Browser-based Combinatorial Library Enumeration
scFv and Custom Antibody Homology Models
GPU-accelerated Protein Modeling and Protein-Protein Docking
Hydrogen Mass Repartitioning for Accelerating MD and TI
Database Viewer SNFG Display, Graphic Objects, and Enhanced Plotting
Integrated Discovery Platform
Multiple Platform Support
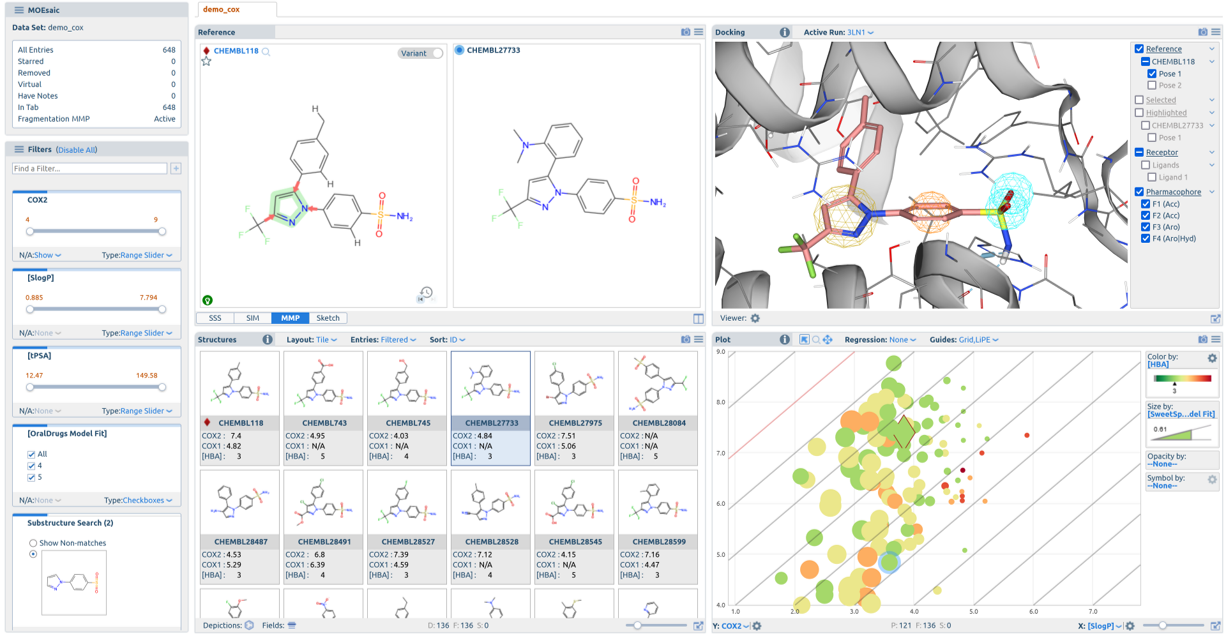
MOEsaic 2022.02
Latest Features
MOEsaic Docking Calculations with Real-Time Visualization of Results
SAR, Matched Molecular Pairs and R-Groups Analysis and Visualization
Property Models for Comparing and Analyzing Compounds
Web-Based SAR Explorer
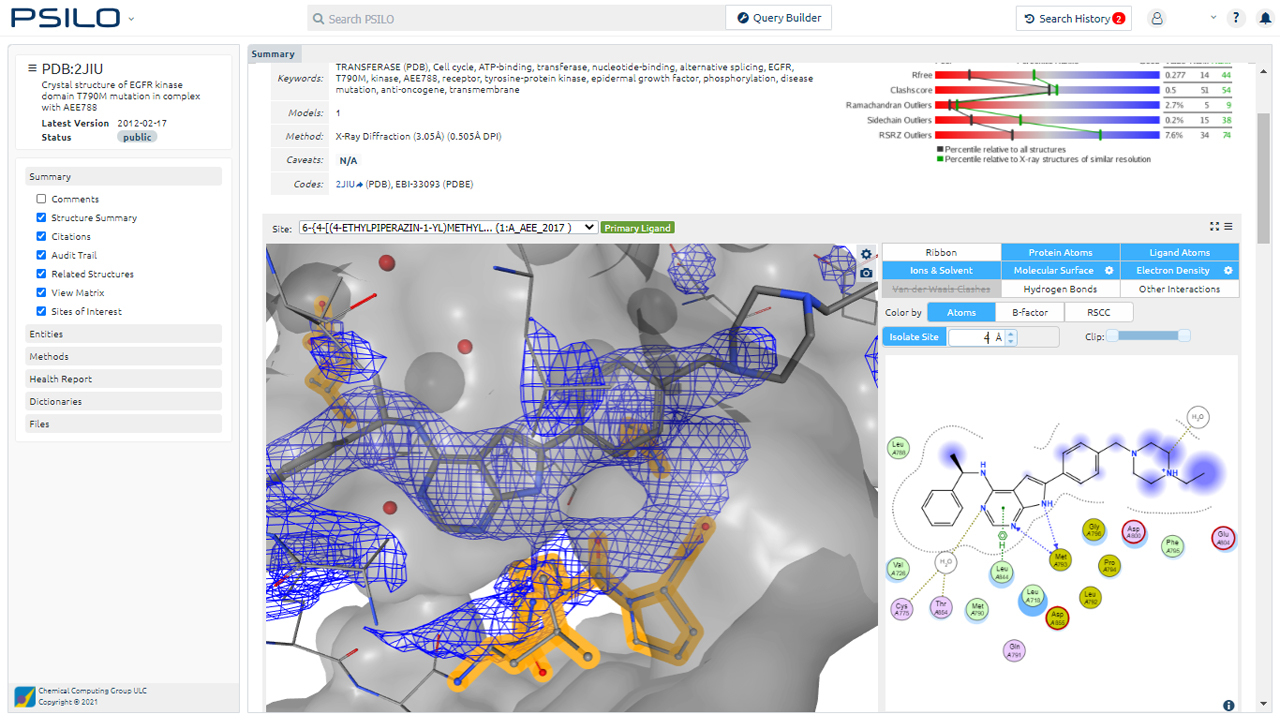
PSILO 2022.1206
Latest Features
Database Federation for Seamless Integration of Public, Private, and AlphaFold PSILO Databases
Streamlined Interface for PSILO Family Analysis and Refinement
Flexible Deposit Validation with Customizable Quality Checks
Broader Support for cryo-EM Maps, Including Public EMDB Maps